FEI TIA software store one TEM image in a pair of files with extension .ser and .emi. The .ser file stores the image pixel information. The data structure of .ser is well documented and there are a number of scripts can be used to read the image info.
However, .emi file is generally not readable outside FEI TIA software. Thanks to the clue from Peter in his blog. The .emi file contains the image metadata (in sort of XML format), log data mixed with a lot of bizzare unreadable code. It is now possible to use the following code to extract the image metadata, i.e. stage position, energy filter setting, etc. It is also possible to use the directory in .emi file pointing to the associated .ser file(s).
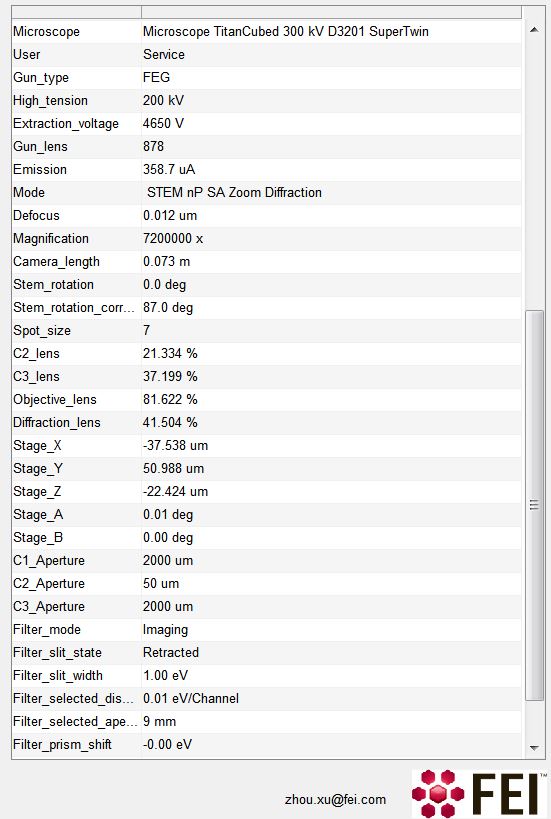
This file contains hidden or bidirectional Unicode text that may be interpreted or compiled differently than what appears below. To review, open the file in an editor that reveals hidden Unicode characters.
Learn more about bidirectional Unicode characters
| % emiReader() function | |
| % Description: | |
| % File reader for .emi files from FEI's TIA programs for electron | |
| % microscopes. Reads in data from .EMI files and the data is | |
| % Parameters: | |
| % fname – the filename of the EMI file to be read. If not provided | |
| % the program opens a dialog box to choose a file | |
| % Output: | |
| % metadata – a matlab structure that contains the microscope | |
| % acquisition information | |
| % Author: | |
| % Zhou Xu 2016-07-06 | |
| % | |
| %———————- NO WARRANTY —————— THIS PROGRAM IS | |
| %PROVIDED AS-IS WITH ABSOLUTELY NO WARRANTY OR GUARANTEE OF ANY KIND, | |
| %EITHER EXPRESSED OR IMPLIED, INCLUDING BUT NOT LIMITED TO, THE IMPLIED | |
| %WARRANTIES OF MERCHANABILITY AND FITNESS FOR A PARTICULAR PURPOSE. IN NO | |
| %EVENT SHALL THE AUTHOR BE LIABLE FOR DAMAGES RESULTING FROM THE USE OR | |
| %INABILITY TO USE THIS PROGRAM (INCLUDING BUT NOT LIMITED TO LOSS OF DATA | |
| %OR DATA BEING RENDERED INACCURATE OR LOSSES SUSTAINED BY YOU OR THIRD | |
| %PARTIES OR A FAILURE OF THE PROGRAM TO OPERATE WITH ANY OTHER PROGRAM). | |
| %———————————————————————— | |
| function [metadata] = emiReader(fName) | |
| fPath = ''; | |
| cellFlag = 0; | |
| if nargin == 0 | |
| [fName, fPath] = uigetfile('*.emi'); | |
| end | |
| if fName == 0 | |
| error('No file opened.'); | |
| end | |
| [FID, FIDmessage] = fopen([fPath fName],'rb'); | |
| if FID == -1 | |
| disp(fName) | |
| error(['Issue opening file: ' FIDmessage]) | |
| end | |
| text = fileread([fPath fName]); | |
| ObjectInfo = text(strfind(text, '<ObjectInfo>'):strfind(text, '</ObjectInfo>')+12); | |
| %%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%% | |
| % find associated .Ser file | |
| SerIndex1 = strfind(text, '<ObjectInfo>')-600; | |
| SerIndex2 = strfind(text, '<ObjectInfo>')-15; | |
| metadata.SerDir = cell(length(SerIndex1),1); | |
| for m = 1: length(SerIndex1) | |
| SerDirRaw = text(SerIndex1(m):SerIndex2(m)); | |
| SerFNameIndex = strfind(SerDirRaw,'\')+1; | |
| metadata.SerFName{m,1} = SerDirRaw(SerFNameIndex(1,length(SerFNameIndex)):strfind(SerDirRaw,'.ser')+3); | |
| metadata.SerDir{m,1} = [fPath metadata.SerFName{m,1}]; | |
| end | |
| % detector range | |
| metadata.Detector_Pixel = [str2num(ObjectInfo(strfind(ObjectInfo, '<DetectorPixelHeight>')+ length('<DetectorPixelHeight>'):strfind(ObjectInfo, '</DetectorPixelHeight>')-1)) str2num(ObjectInfo(strfind(ObjectInfo, '<DetectorPixelWidth>')+ length('<DetectorPixelWidth>'):strfind(ObjectInfo, '</DetectorPixelWidth>')-1))]; | |
| DetectorRangeX1 = str2num( ObjectInfo(strfind(ObjectInfo, '<StartX>')+ length('<StartX>'):strfind(ObjectInfo, '</StartX>')-1) ); | |
| DetectorRangeX2 = str2num( ObjectInfo(strfind(ObjectInfo, '<EndX>')+ length('<EndX>'):strfind(ObjectInfo, '</EndX>')-1) ); | |
| DetectorRangeY1 = str2num( ObjectInfo(strfind(ObjectInfo, '<StartY>')+ length('<StartY>'):strfind(ObjectInfo, '</StartY>')-1) ); | |
| DetectorRangeY2 = str2num( ObjectInfo(strfind(ObjectInfo, '<EndY>')+ length('<EndY>'):strfind(ObjectInfo, '</EndY>')-1) ); | |
| metadata.Detector_Range = [DetectorRangeX1 DetectorRangeX2; DetectorRangeY1 DetectorRangeY2 ]; | |
| %%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%% | |
| % Uuid | |
| metadata.Uuid = ObjectInfo(strfind(ObjectInfo, '<Uuid>')+ length('<Uuid>'):strfind(ObjectInfo, '</Uuid>')-1); | |
| % file directory | |
| metadata.File = [fPath fName]; | |
| % Experimental Conditions/Microscope Conditions | |
| metadata.EXPERIMENTAL_CONDITIONS = ''; % title | |
| Microscope_Condition = ObjectInfo(strfind(ObjectInfo, '<MicroscopeCondition>')+ length('<MicroscopeCondition>'):strfind(ObjectInfo, '</MicroscopeCondition>')-1); | |
| metadata.Accelerating_Voltage = ObjectInfo(strfind(ObjectInfo, '<AcceleratingVoltage>')+ length('<AcceleratingVoltage>'):strfind(ObjectInfo, '</AcceleratingVoltage>')-1); | |
| metadata.Tilt1 = ObjectInfo(strfind(ObjectInfo, '<Tilt1>')+ length('<Tilt1>'):strfind(ObjectInfo, '</Tilt1>')-1); | |
| metadata.Tilt2 = ObjectInfo(strfind(ObjectInfo, '<Tilt2>')+ length('<Tilt2>'):strfind(ObjectInfo, '</Tilt2>')-1); | |
| %%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%% | |
| % Acquire Info | |
| %%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%%% | |
| metadata.ACQUISITION = ''; % title | |
| metadata.Manufacturer = ObjectInfo(strfind(ObjectInfo, '<Manufacturer>')+ length('<Manufacturer>'):strfind(ObjectInfo, '</Manufacturer>')-1); | |
| % metadata.Acquire_Date = ObjectInfo(strfind(ObjectInfo, '<AcquireDate>')+ length('<AcquireDate>'):strfind(ObjectInfo, '</AcquireDate>')-1); | |
| Acquire_Date = ObjectInfo(strfind(ObjectInfo, '<AcquireDate>')+ length('<AcquireDate>'):strfind(ObjectInfo, '</AcquireDate>')-1); | |
| Acquire_Date = datetime(Acquire_Date,'InputFormat','eee MMM dd HH:mm:ss yyyy'); % 'Tue Apr 05 14:20:17 2016' | |
| metadata.Acquire_Date = datestr(Acquire_Date, 'yyyy-mm-dd HH:MM:SS.FFF'); | |
| mode = strfind(ObjectInfo, 'STEM'); | |
| % Dwell_Time_Path | |
| if isempty(mode)==1 | |
| metadata.Camera_Name = ObjectInfo(strfind(ObjectInfo, '<CameraNamePath>')+ length('<CameraNamePath>'):strfind(ObjectInfo, '</CameraNamePath>')-1); | |
| metadata.Integration_Time = ObjectInfo(strfind(ObjectInfo, '<DwellTimePath>')+ length('<DwellTimePath>'):strfind(ObjectInfo, '</DwellTimePath>')-1); | |
| metadata.Binning = ObjectInfo(strfind(ObjectInfo, '<Binning>')+ length('<Binning>'):strfind(ObjectInfo, '</Binning>')-1); | |
| else | |
| metadata.Dwell_Time = ObjectInfo(strfind(ObjectInfo, '<DwellTimePath>')+ length('<DwellTimePath>'):strfind(ObjectInfo, '</DwellTimePath>')-1); | |
| metadata.Frame_Time = ObjectInfo(strfind(ObjectInfo, '<FrameTime>')+ length('<FrameTime>'):strfind(ObjectInfo, '</FrameTime>')-1); | |
| end | |
| % detector range | |
| metadata.Detector_Range_X = [num2str(DetectorRangeX1) ' to ' num2str(DetectorRangeX2)]; | |
| metadata.Detector_Range_Y = [num2str(DetectorRangeY1) ' to ' num2str(DetectorRangeY2)]; | |
| % ExperiementalDescription | |
| metadata.MICROSCOPE_INFO = ''; % title | |
| ExperimentalDescription = ObjectInfo(strfind(ObjectInfo, '<ExperimentalDescription>')+ length('<ExperimentalDescription>'):strfind(ObjectInfo, '</ExperimentalDescription>')-1); | |
| DataIndex1 = strfind(ExperimentalDescription, '<Data>')+length('<Data>'); | |
| DataIndex2 = strfind(ExperimentalDescription, '</Data>')-1; | |
| Data = cell(length(DataIndex1),1); | |
| Field = cell(length(DataIndex1),1); | |
| Value = cell(length(DataIndex1),1); | |
| Unit = cell(length(DataIndex1),1); | |
| ValueUnit = cell(length(DataIndex1),1); | |
| for i = 1: length(DataIndex1) | |
| Data{i,1} = ExperimentalDescription(DataIndex1(i):DataIndex2(i)); | |
| temp = Data{i}; | |
| Field{i,1} = temp(strfind(temp, '<Label>')+length('<Label>'):strfind(temp, '</Label>')-1); | |
| Field{i,1}(Field{i,1}==' ') = '_'; | |
| Value{i,1} = temp(strfind(temp, '<Value>')+ length('<Value>'):strfind(temp, '</Value>')-1); | |
| Unit{i,1} = temp(strfind(temp, '<Unit>')+ length('<Unit>'):strfind(temp, '</Unit>')-1); | |
| ValueUnit{i,1} = [Value{i,1} ' ' Unit{i,1}]; | |
| metadata.(Field{i,1}) = ValueUnit{i,1}; | |
| end | |
| end |

Leave a comment